DICOM File Processing
We’ll explore some open source libraries in different programming languages and how you can use them to process DICOM files. We’ll cover the basics of removing, modifying, adding data elements, changing compression, creating DICOM files for testing, and masking parts of images for de-identification purposes.
If you are new to the DICOM Standard and you are not sure how the DICOM file format works, please read DICOM Basics first.
Disclaimer: Everything presented here is part of public knowledge and can be found in referenced material.
Open Source Libraries
The following are some of the popular open source libraries for processing DICOM files:
- PixelMed - Java DICOM Toolkit which is a stand-alone DICOM toolkit that implements code for reading and creating DICOM data, DICOM network and file support, support for display of images, reports, and much much more
- pydicom - Pure Python package for working with DICOM files. It lets you read, modify and write DICOM data in an easy “pythonic” way.
- Grassroots DICOM (GDCM) - Cross-platform library written in C++ for DICOM medical files. It is automatically wrapped to Python/C#/Java and PHP which allows you to use the language you are familiar with and to integrate it with other applications.
- dicomParser - Lightweight library for parsing DICOM in modern HTML5 based web browsers. dicomParser is fast, easy to use and has no required external dependencies.
- DCMTK - is a collection of libraries and applications implementing large parts the DICOM standard. It includes software for examining, constructing and converting DICOM image files, handling offline media, sending and receiving images over a network connection, as well as demonstrative image storage and worklist servers. DCMTK is written in a mixture of ANSI C and C++.
The choice depends on your use case and, of course, on the language you are comfortable with.
These libraries implement most of the things from the DICOM Standard and there is no way I can cover all of them in this article. Therefore, we will focus only on the PixelMed, pydicom, some of GDCM tools, and switch between them to implement different functionalities.
Setup
PixelMed
PixelMed is written in Java, so you’ll need Java 1.8 or higher. After you
install Java, go to PixelMed Directory Tree,
select the current edition, and download the pixelmed.jar (or use Maven/Gradle
if you are familiar with them).
If you are not sure how to use this jar, I suggest you download
Eclipse IDE,
create a Java project and add pixelmed.jar to the project, see:
How to import a jar in Eclipse.
PixelMed’s API documentation can be found at PixelMed JavaDocs.
GDCM
Source code: Grassroots DICOM.
There are premade tools which you can use:
- gdcmdump - dumps a DICOM file, it will display the structure and values contained in the specified DICOM file.
- gdcmanon - tool to anonymize a DICOM file.
- gdcmdiff - dumps differences of two DICOM files
- gdcmconv - tool to convert DICOM to DICOM etc
To use them you can either compile the source or download the binaries from GDCM Releases.
If you are on Linux, you’ll have to add the gdcm
libfolder to/etc/ld.so.conffile or create new.confin folder.d. Then runsudo ldconfig. In order to have gdcm applications available in the terminal as commands you’ll have to addbinto$PATH(globally or in~/.bash_profile).
pydicom
This library requires python >= 3.6.1. I suggest you set up a virtual environment where you can install packages using a dependency manager (poetry, pipenv, or any other).
To get familiar with the library, please see: pydicom documentation.
DICOM
Exploring Structure
GDCM
To explore the structure of a DICOM file, we can use gdcmdump:
gdcmdump <path-to-dicom-file>
This will give us an output like:
# Dicom-File-Format
# Dicom-Meta-Information-Header
# Used TransferSyntax:
(0002,0000) UL 194 # 4,1 File Meta Information Group Length
(0002,0001) OB 00\01 # 2,1 File Meta Information Version
(0002,0002) UI [1.2.840.10008.5.1.4.1.1.12.2] # 28,1 Media Storage SOP Class UID
(0002,0003) UI [1.3.6.1.4.1.5962.1.1.0.0.0.1168612284.20369.0.3] # 48,1 Media Storage SOP Instance UID
(0002,0010) UI [1.2.840.10008.1.2.1] # 20,1 Transfer Syntax UID
(0002,0012) UI [1.3.6.1.4.1.5962.2] # 18,1 Implementation Class UID
(0002,0013) SH [DCTOOL100 ] # 10,1 Implementation Version Name
(0002,0016) AE [CLUNIE1 ] # 8,1 Source Application Entity Title
# Dicom-Data-Set
# Used TransferSyntax: 1.2.840.10008.1.2.1
(0008,0005) CS [ISO_IR 100] # 10,1-n Specific Character Set
(0008,0008) CS [ORIGINAL\PRIMARY\SINGLE PLANE ] # 30,2-n Image Type
(0008,0012) DA [20070112] # 8,1 Instance Creation Date
(0008,0013) TM [093126] # 6,1 Instance Creation Time
(0008,0014) UI [1.3.6.1.4.1.5962.3] # 18,1 Instance Creator UID
(0008,0016) UI [1.2.840.10008.5.1.4.1.1.12.2] # 28,1 SOP Class UID
(0008,0018) UI [1.3.6.1.4.1.5962.1.1.0.0.0.1168612284.20369.0.3] # 48,1 SOP Instance UID
...
This tool offers a quick and easy way to explore DICOM files and debug applications that process DICOM files.
Looking at the output we can see the Dicom-Meta-Information-Header and
Dicom-Data-Set. Additionally, for each Data Element, we have a Tag
(gggg,eeee) then a VR, followed by a Value, and the information after #
represents: VL, VM, and Tag Name.
gdcmdump comes with a lot of options that you can use to generate an output,
for more information please see: gdcmdump
PixelMed
The AttributeList class is a class in the PixelMed that maintains a list of
individual DICOM attributes. It could be used to get the structure of a file,
modify it, and save it.
Using .read(java.lang.String name) we can read the tags:
package pixelmed_demo;
import java.io.IOException;
import com.pixelmed.dicom.AttributeList;
import com.pixelmed.dicom.DicomException;
public class demo_main {
public static void main(String[] args) {
String filepath = args[0];
AttributeList attList = new AttributeList();
try {
attList.read(filepath);
System.out.println(attList.toString());
} catch (IOException | DicomException e) {
System.out.println("Oops! Error: " + e.getMessage());
}
}
}
Passing the same DICOM file we used in gdcmdump example as an argument,
we get:
(0x0002,0x0000) FileMetaInformationGroupLength VR=<UL> VL=<0x4> [0xc2]
(0x0002,0x0001) FileMetaInformationVersion VR=<OB> VL=<0x2> []
(0x0002,0x0002) MediaStorageSOPClassUID VR=<UI> VL=<0x1c> <1.2.840.10008.5.1.4.1.1.12.2>
(0x0002,0x0003) MediaStorageSOPInstanceUID VR=<UI> VL=<0x30> <1.3.6.1.4.1.5962.1.1.0.0.0.1168612284.20369.0.3>
(0x0002,0x0010) TransferSyntaxUID VR=<UI> VL=<0x14> <1.2.840.10008.1.2.1>
(0x0002,0x0012) ImplementationClassUID VR=<UI> VL=<0x12> <1.3.6.1.4.1.5962.2>
(0x0002,0x0013) ImplementationVersionName VR=<SH> VL=<0xa> <DCTOOL100 >
(0x0002,0x0016) SourceApplicationEntityTitle VR=<AE> VL=<0x8> <CLUNIE1 >
(0x0008,0x0005) SpecificCharacterSet VR=<CS> VL=<0xa> <ISO_IR 100>
(0x0008,0x0008) ImageType VR=<CS> VL=<0x1e> <ORIGINAL\PRIMARY\SINGLE PLANE >
(0x0008,0x0012) InstanceCreationDate VR=<DA> VL=<0x8> <20070112>
(0x0008,0x0013) InstanceCreationTime VR=<TM> VL=<0x6> <093126>
(0x0008,0x0014) InstanceCreatorUID VR=<UI> VL=<0x12> <1.3.6.1.4.1.5962.3>
(0x0008,0x0016) SOPClassUID VR=<UI> VL=<0x1c> <1.2.840.10008.5.1.4.1.1.12.2>
(0x0008,0x0018) SOPInstanceUID VR=<UI> VL=<0x30> <1.3.6.1.4.1.5962.1.1.0.0.0.1168612284.20369.0.3>
...
which gives the same output as gdcmdump but in a different format.
Remove Data Element
Let’s try to remove the InstanceCreatorUID (0008,0014) from the Data Set and
save the DICOM object as a new file.
PixelMed
The AttributeList has a lot of options for manipulating Data Sets,
we can remove a whole group, all private tags, specific tag etc.
package pixelmed_demo;
import java.io.IOException;
import com.pixelmed.dicom.AttributeList;
import com.pixelmed.dicom.AttributeTag;
import com.pixelmed.dicom.Attribute;
import com.pixelmed.dicom.DicomException;
public class demo_main {
public static void main(String[] args) {
String filepath = args[0];
AttributeList attList = new AttributeList();
AttributeTag instanceCreatorTag = new AttributeTag(0x0008, 0x0014);
AttributeTag transferSyntaxTag = new AttributeTag(0x0002, 0x0010);
try {
// Read DICOM
attList.read(filepath);
// Get TransferSyntaxUID
Attribute transferSyntaxAtt = attList.get(transferSyntaxTag);
String transferSyntaxUID = transferSyntaxAtt.getSingleStringValueOrEmptyString();
// Remove Tag
attList.remove(instanceCreatorTag);
// Write DICOM
attList.write("test.dcm", transferSyntaxUID, true, true);
} catch (IOException | DicomException e) {
System.out.println("Oops! Error: " + e.getMessage());
}
}
}
This will output a new DICOM file test.dcm which doesn’t contain the
InstanceCreatorUID.
To confirm this we can run gdcmdiff which dumps the difference between two DICOM files:
gdcmdiff <path_to_new_file> <path_to_old_file>
The output is:
(0008,0014) UI [only file 2] [1.3.6.1.4.1.5962.3] # Instance Creator UID
-------------
as expected.
You may notice that as we deal with many attributes, defining a specific tag as:
AttributeTag transferSyntaxTag = new AttributeTag(0x0002, 0x0010);
becomes tedious. Fortunately, there are better ways. One of them is to use the
TagFromName which contains constants that map names to tags, and the other
is to use the DicomDictionary to do a lookup:
// Using TagFromName
AttributeTag transferSyntaxTag = TagFromName.TransferSyntaxUID;
// Using DicomDictionary
AttributeTag transferSyntaxTag = DicomDictionary.StandardDictionary.getTagFromName("TransferSyntaxUID")
pydicom
Let’s do the same using the pydicom. To do this, we will use the dcmread to
read a DICOM file which returns a FileDataset instance that we can edit and
then save.
from pydicom import dcmread
if __name__ == "__main__":
with open("<path-to-dicom-file>", "rb") as f_in:
ds = dcmread(f_in)
del ds.InstanceCreatorUID
ds.save_as("test.dcm")
Modify/Add Data Element
PixelMed
To modify/add a Data Element to the Data Set, we create an AttributeList
instance and an Attribute instance, then put the attribute to the list:
package pixelmed_demo;
import java.io.IOException;
import com.pixelmed.dicom.AttributeList;
import com.pixelmed.dicom.AttributeTag;
import com.pixelmed.dicom.Attribute;
import com.pixelmed.dicom.DicomException;
import com.pixelmed.dicom.TagFromName;
import com.pixelmed.dicom.PersonNameAttribute;
import com.pixelmed.dicom.CodeStringAttribute;
public class demo_main {
public static void main(String[] args) {
String filepath = args[0];
AttributeList attList = new AttributeList();
try {
// Read DICOM
attList.read(filepath);
// Get TransferSyntaxUID
Attribute transferSyntaxAtt = attList.get(TagFromName.TransferSyntaxUID);
String transferSyntaxUID = transferSyntaxAtt.getSingleStringValueOrEmptyString();
// Modify Existing Tag
Attribute patientName = new PersonNameAttribute(TagFromName.PatientName);
patientName.addValue("TestName");
attList.put(TagFromName.PatientName, patientName);
// Add New Tag
Attribute newAtt = new CodeStringAttribute(new AttributeTag(0x0011, 0x0010));
newAtt.addValue("SomeRandomString");
attList.put(newAtt);
// Write DICOM
attList.write("test.dcm", transferSyntaxUID, true, true);
} catch (IOException | DicomException e) {
System.out.println("Oops! Error: " + e.getMessage());
}
}
}
This will create a new DICOM file with modified PatientName tag and a new tag
(0011,0010). If we use gdcmdiff to check the differences between the old
and the new file, we get exactly what we expect:
(0010,0010) PN [from file 1] [TestName] # Patient's Name
(0010,0010) PN [from file 2] [Test^FluroWithDisplayShutter] # Patient's Name
-------------
(0011,0010) CS [only file 1] [SomeRandomString] # Private Creator
-------------
pydicom
To do the same in python:
from pydicom import dcmread
if __name__ == "__main__":
with open("<path-to-dicom-file>", "rb") as f_in:
ds = dcmread(f_in)
# Modify Existing Tag
ds.PatientName = "TestName"
# Add New Tag
ds.add_new([0x0011, 0x0010], "CS", "SomeRandomString")
# Write DICOM
ds.save_as("test.dcm")
which gives use the same result.
If we want to add a tag from the DICOM Standard but we are not sure about its
VR, we can use the dictionary_VR:
>>> from pydicom.datadict import dictionary_VR
>>> dictionary_VR([0x0028, 0x1050])
'DS'
Add Nested Data Element
Let’s add a private nested data element that contains two items which contain the same private attributes but with different values. The process is the same for the tags from the DICOM Standard.
PixelMed
To add a nested tag using the PixelMed, we have to define a SequenceAttribute.
This attribute will contain Sequence Items which we create using the
AttributeList. After adding Data Elements to the AttributeList we add the list
to the SequenceAttribute using addItem(AttributeList item):
package pixelmed_demo;
import java.io.IOException;
import com.pixelmed.dicom.AttributeList;
import com.pixelmed.dicom.AttributeTag;
import com.pixelmed.dicom.Attribute;
import com.pixelmed.dicom.DicomException;
import com.pixelmed.dicom.TagFromName;
import com.pixelmed.dicom.SequenceAttribute;
import com.pixelmed.dicom.CodeStringAttribute;
import com.pixelmed.dicom.DateAttribute;
public class demo_main {
public static void main(String[] args) {
String filepath = args[0];
AttributeList attList = new AttributeList();
try {
// Read DICOM
attList.read(filepath);
// Get TransferSyntaxUID
Attribute transferSyntaxAtt = attList.get(TagFromName.TransferSyntaxUID);
String transferSyntaxUID = transferSyntaxAtt.getSingleStringValueOrEmptyString();
// Sequence Attribute
SequenceAttribute seq = new SequenceAttribute(new AttributeTag(0x0011, 0x0010));
// Sequence Item 1
AttributeList seqItemOne = new AttributeList();
Attribute attSeqOneOne = new CodeStringAttribute(new AttributeTag(0x0011, 0x0100));
attSeqOneOne.addValue("Sequence One Value");
Attribute attSeqOneTwo = new DateAttribute(new AttributeTag(0x0011, 0x0102));
attSeqOneTwo.addValue("2022-01-01");
seqItemOne.put(attSeqOneOne);
seqItemOne.put(attSeqOneTwo);
// Sequence Item 2
AttributeList seqItemTwo = new AttributeList();
Attribute attSeqTwoOne = new CodeStringAttribute(new AttributeTag(0x0011, 0x0100));
attSeqTwoOne.addValue("Sequence Two Value");
Attribute attSeqTwoTwo = new DateAttribute(new AttributeTag(0x0011, 0x0102));
attSeqTwoTwo.addValue("2022-01-02");
seqItemTwo.put(attSeqTwoOne);
seqItemTwo.put(attSeqTwoTwo);
// Add Sequence Items
seq.addItem(seqItemOne);
seq.addItem(seqItemTwo);
attList.put(seq);
// Write DICOM
attList.write("test.dcm", transferSyntaxUID, true, true);
} catch (IOException | DicomException e) {
System.out.println("Oops! Error: " + e.getMessage());
}
}
}
Each sequence item is a Data Set of its own and for each
item we create an AttributeList instance where we add attributes, in the
end we add these lists as sequence items to a sequence attribute.
If we check the result with gdcmdump, we should see:
(0011,0010) SQ (LO) (Sequence with undefined length) # u/l,1 Private Creator
(fffe,e000) na (Item with undefined length)
(0011,0100) CS [Sequence One Value] # 18,? (1) Private Element With Empty Private Creator
(0011,0102) DA [2022-01-01] # 10,? (1) Private Element With Empty Private Creator
(fffe,e00d)
(fffe,e000) na (Item with undefined length)
(0011,0100) CS [Sequence Two Value] # 18,? (1) Private Element With Empty Private Creator
(0011,0102) DA [2022-01-02] # 10,? (1) Private Element With Empty Private Creator
(fffe,e00d)
(fffe,e0dd)
which is what we wanted.
pydicom
The same can be done using the pydicom, here we define a seq variable that is
just a list of Dataset instances, these data sets have the same meaning as
AttributeList in the PixelMed. After that we access individual data sets
i.e. sequence items and add attributes to them:
from pydicom import dcmread, Dataset
if __name__ == "__main__":
with open("test-dicom-files/image-2.dcm", "rb") as f_in:
ds = dcmread(f_in)
# Sequence Attribute
seq = [Dataset(), Dataset()]
# Sequence Item 1
seq[0].add_new([0x0011, 0x0100], "CS", "Sequence One Value")
seq[0].add_new([0x0011, 0x0102], "DA", "2021-01-01")
# Sequence Item 2
seq[1].add_new([0x0011, 0x0100], "CS", "Sequence Two Value")
seq[1].add_new([0x0011, 0x0102], "DA", "2021-01-02")
# Add Sequence Attribute
ds.add_new([0x0011, 0x0010], 'SQ', seq)
# Write DICOM
ds.save_as("test.dcm")
The result is the same as above.
Change Transfer Syntax
Let’s first change Explicit to Implicit.
Of course, before writing a new DICOM file, we have to update the information about new TransferSyntaxUID and SourceApplicationEntityTitle.
package pixelmed_demo;
import java.io.IOException;
import com.pixelmed.dicom.AttributeList;
import com.pixelmed.dicom.DicomException;
import com.pixelmed.dicom.TransferSyntax;
import com.pixelmed.dicom.FileMetaInformation;
public class demo_main {
public static void main(String[] args) {
String filepath = args[0];
AttributeList attList = new AttributeList();
try {
// Read DICOM
attList.read(filepath);
// Update File Meta Information Header
FileMetaInformation.addFileMetaInformation(attList, TransferSyntax.ImplicitVRLittleEndian, "DicomPlayGround");
// Write DICOM
attList.write("test.dcm", TransferSyntax.ImplicitVRLittleEndian, true, true);
} catch (IOException | DicomException e) {
System.out.println("Oops! Error: " + e.getMessage());
}
}
}
The new file looks like:
# Dicom-File-Format
# Dicom-Meta-Information-Header
# Used TransferSyntax:
(0002,0000) UL 210 # 4,1 File Meta Information Group Length
(0002,0001) OB 00\01 # 2,1 File Meta Information Version
(0002,0002) UI [1.2.840.10008.5.1.4.1.1.12.2] # 28,1 Media Storage SOP Class UID
(0002,0003) UI [1.3.6.1.4.1.5962.1.1.0.0.0.1168612284.20369.0.3] # 48,1 Media Storage SOP Instance UID
(0002,0010) UI [1.2.840.10008.1.2] # 18,1 Transfer Syntax UID
(0002,0012) UI [1.3.6.1.4.1.5962.99.2] # 22,1 Implementation Class UID
(0002,0013) SH [PIXELMEDJAVA001 ] # 16,1 Implementation Version Name
(0002,0016) AE [DicomPlayGround ] # 16,1 Source Application Entity Title
# Dicom-Data-Set
# Used TransferSyntax: 1.2.840.10008.1.2
(0008,0005) ?? (CS) [ISO_IR 100] # 10,1-n Specific Character Set
(0008,0008) ?? (CS) [ORIGINAL\PRIMARY\SINGLE PLANE ] # 30,2-n Image Type
(0008,0012) ?? (DA) [20070112] # 8,1 Instance Creation Date
(0008,0013) ?? (TM) [093126] # 6,1 Instance Creation Time
(0008,0014) ?? (UI) [1.3.6.1.4.1.5962.3] # 18,1 Instance Creator UID
(0008,0016) ?? (UI) [1.2.840.10008.5.1.4.1.1.12.2] # 28,1 SOP Class UID
Compare this to the same file in the Explicit format, the changes are obvious and expected.
However, it’s not so simple if we have compressed pixel data. Depending on the compression and wanted output it could be hard to find needed libraries that can do the conversion.
The PixelMed relies on Java libraries for compressing and decompressing pixel data.
You could use imageIO, the PixelMed’s stand-alone Java JPEG Selective Block
Redaction Codec and Lossless JPEG Decoder, or any other library, but as far
as I know, things can get complicated and inefficient for certain
TransferSyntaxUIDs i.e. compression algorithms.
In my opinion, when it comes to compression, the better option is to use GDCM only or GDCM in combination with the pydicom. To get the better understanding of supported transfer syntaxes, please see the table at Supported Transfer Syntaxes.
For now, we can use gdcmconv tool to play with different transfer syntaxes. There are a lot of options, and you can explore them at gdcmconv.
To convert to JPEG Lossless i.e. 1.2.840.10008.1.2.4.70 we can use:
gdcmconv --jpeg -i <input-file> -o <output-file>
If your input file was uncompressed, you should see a significant loss in size of the output file.
Create DICOM from an Image
Creating a DICOM file from an image can be really useful in many cases. Especially, when it comes to testing:
- Application should be tested in test/dev environment where you cannot use real world scans
- De-identification process that masks certain parts of images should be tested for different image resolutions etc.
To achieve this you can use the ImageToDicom:
package pixelmed_demo;
import java.io.IOException;
import com.pixelmed.dicom.ImageToDicom;
import com.pixelmed.dicom.DicomException;
public class demo_main {
public static void main(String[] args) {
String filepath = args[0];
try {
new ImageToDicom(
filepath,
"test.dcm", // Output path
"Vladsiv", // Patient Name
"TEST-98341-VladSiv", // Patient ID
"847542", // Study ID
"1", // Series Number
"1", // Instance Number
"US", // Modality
"1.2.840.10008.5.1.4.1.1.6.1" // SOP Class UID
);
} catch (IOException | DicomException e) {
System.out.println("Oops! Error: " + e.getMessage());
}
}
}
The result is:
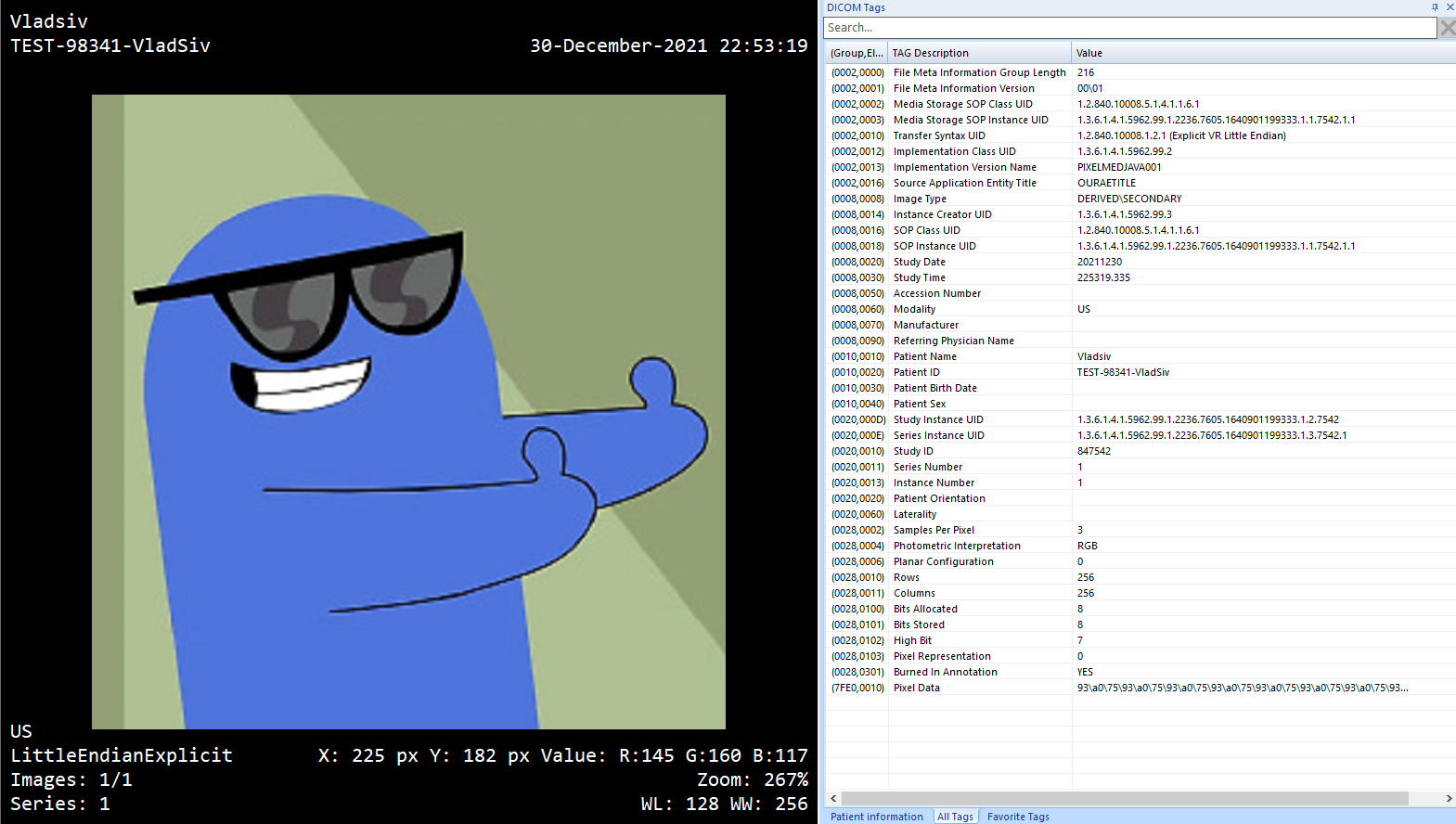
Of course, you can then process the output DICOM file, add/modify data elements, and tailor the file for your specific needs.
To do the same using the pydicom, please see this thread: PNG to DICOM with pydicom
Blackout Image
In certain circumstances, we want to blackout certain parts of DICOM images. This is usually done to remove the Protected Health Information - PHI that can be found on an image.
To do this using the PixelMed we can use the ImageEditUtilities and the method:
blackout(SourceImage srcImg, AttributeList list, java.util.Vector shapes):
package pixelmed_demo;
import java.awt.Shape;
import java.awt.Rectangle;
import java.util.Vector;
import com.pixelmed.display.ImageEditUtilities;
import com.pixelmed.display.SourceImage;
import com.pixelmed.dicom.AttributeList;
import com.pixelmed.dicom.Attribute;
import com.pixelmed.dicom.TagFromName;
public class demo_main {
public static void main(String[] args) {
String filepath = args[0];
AttributeList attList = new AttributeList();
try {
// Read DICOM file
attList.read(filepath);
// Get Transfer Syntax
Attribute transferSyntaxAtt = attList.get(TagFromName.TransferSyntaxUID);
String transferSyntaxUID = transferSyntaxAtt.getSingleStringValueOrEmptyString();
// Create Area to blackout
Vector<Shape> shapes = new Vector<Shape>();
Shape shape = new Rectangle(35, 60, 140, 50);
shapes.add(shape);
// Define Image and Perform Blackout
SourceImage sImg = new SourceImage(attList);
ImageEditUtilities.blackout(sImg, attList, shapes);
// Write DICOM
attList.write("test.dcm", transferSyntaxUID, true, true);
} catch (Exception e) {
System.out.println("Oops! Error: " + e.getMessage());
}
}
}
This will give us:
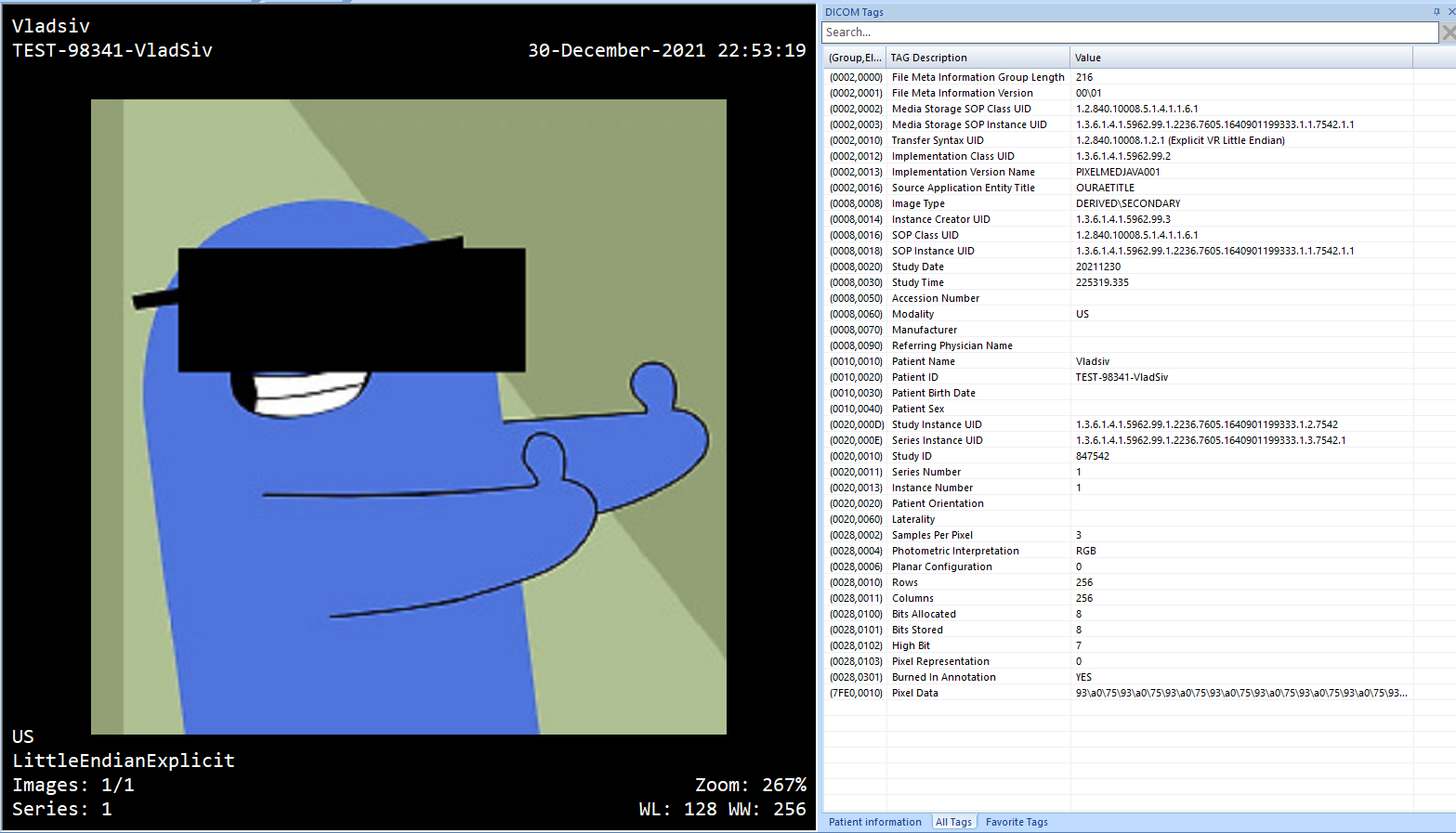
Final Words
This article gives a brief introduction to basics of processing DICOM files using some of the open source libraries.
There are many awesome libraries and DICOM applications that I didn’t mention and I encourage you to go through provided material and play with them on your own.
I hope this helps you get a better understanding how you can process DICOM files and start building your own DICOM applications.
If you have any questions or suggestions, please reach out, I’m always available.
Leave a comment